Model Overview & Architecture#
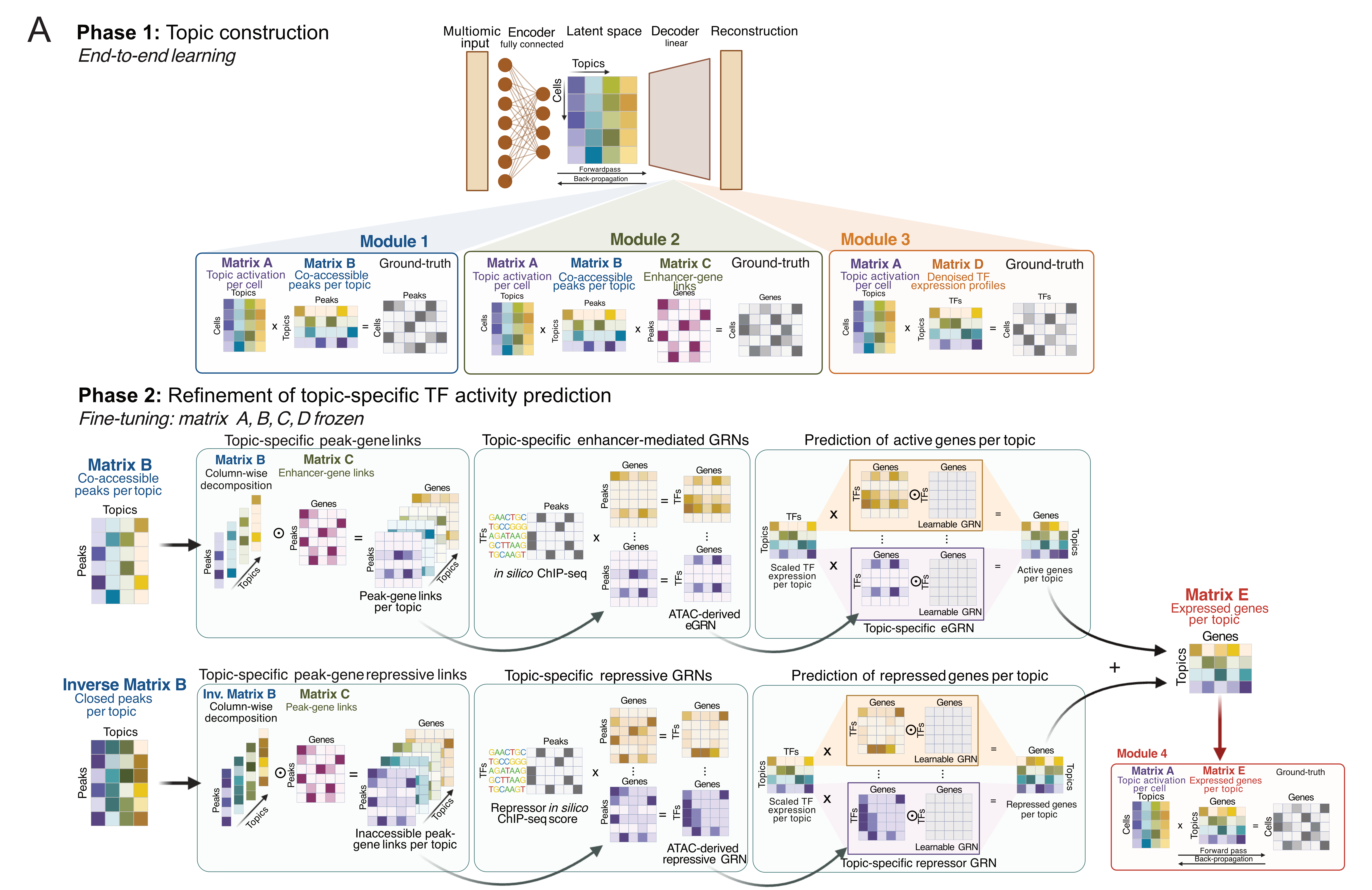
scDoRI (single-cell Deep Multi-Omic Regulatory Inference) is a computational framework that jointly models paired single-cell RNA-seq and ATAC-seq profiles to infer enhancer-mediated gene regulatory networks (eGRNs). Unlike existing pipelines that treat dimensionality reduction and regulatory inference as distinct modules, scDoRI unifies them in a single encoder–decoder architecture grounded in biological priors.
At its core, the model learns topics – regulatory modules that link co-accessible chromatin regions, their cis-mediated target genes, and upstream activator and repressor transcription factors (TFs). Each cell is represented as a probabilistic mixture over topics, allowing for a continuous and interpretable view of transcriptional regulation.
Architectural Components#
scDoRI consists of two primary components:
Encoder#
Projects high-dimensional RNA and ATAC profiles into a shared latent topic space.
Comprised of parallel neural networks (one each for RNA and ATAC), with outputs concatenated and mapped into topic logits.
Final output is a topic mixture vector for each cell, constrained via a softmax activation (topics sum up to 1 per cell).
Decoder#
The decoder reconstructs observed data modalities from the shared latent topic space, enforcing biologically constrained logic through four modules:
Module 1: ATAC Reconstruction
Reconstructs peak accessibility using a topic–peak weight matrix.
Includes batch-specific offsets to account for experimental variability.
Applies L1 regularization on topic–peak weights to encourage sparsity for interpretability.
Module 2: RNA-from-ATAC Prediction
Reconstructs gene expression based on predicted chromatin accessibility from Module 1.
Employs a learnable gene–peak linkage matrix, constrained by genomic proximity (e.g., peaks within 150kb of the gene).
Module 3: TF Expression Reconstruction
Learns topic-to-TF expression mappings, allowing the latent space to capture transcription factor expression programs.
Module 4: Signed TF–Gene Network Inference
Computes signed topic-specific TF–gene links by integrating:
Precomputed TF–peak motif scores (activators and repressors)
Topic-wise chromatin accessibility
Gene–peak associations
Topic-level TF expression
Refines scores using a learnable 3D TF–gene–topic matrix.
Produces a final signed GRN, used to reconstruct gene expression from TF expression per topic (Module 3).
Training Phases#
Phase 1: Topic Construction#
The encoder and Modules 1–3 are trained jointly using multimodal reconstruction losses.
Peak accessibility, gene expression, and TF expression are reconstructed from latent topics.
Objective functions include Poisson likelihood loss (ATAC) and Negative Binomial likelihood loss (RNA), with regularization to promote sparsity and interpretability.
Phase 2: GRN Refinement#
Module 4 is introduced to learn topic-specific GRNs from chromatin context, peak - gene links and TF- peak links (from insilico-ChIP-seq, introduced in https://www.biorxiv.org/content/10.1101/2022.06.15.496239v1).
Adds trainable activator/repressor TF–gene link matrices per topic.
Predicts RNA from TF expression using inferred GRNs.
Earlier modules can optionally be frozen to preserve previously learned topic representations.
Schematic Overview#
The figure above provides a schematic overview of the scDoRI model. Modules are color-coded by training phase, and matrix roles are explicitly annotated. Phase 1 involves joint training of the encoder and Modules 1–3. Phase 2 fine-tunes Module 4 to enable topic-specific GRN inference.